De novo transcriptome sequencing reveals candidate genes involved in orange shell coloration of bay scallop Argopecten irradians*
TENG Wen (騰文) CONG Rihao (叢日浩) QUE Huayong (闕華勇) ZHANG Guofan (張國范)
1 Institute of Oceanology, Chinese Academy of Sciences, Qingdao 266071, China
2 University of Chinese Academy of Sciences, Beijing 100049, China
3 Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao 266071, China
4 National & Local Joint Engineering Laboratory of Ecological Mariculture, Qingdao 266000, China
Abstrac t Molluscan shell color has received persistent attention for its distinctive diversity and complexity.In the present study, six transcriptome libraries obtained from two developmental stages, pre-pigmentation and post-pigmentation, were used for paired-end sequencing in the bay scallop Argopecten irradians. In total, 289 839 646 paired-end reads were assembled into 70 929 transcripts. Using BLASTX and BLASTN,30 896 unigenes were successfully annotated against the SWISS-PROT, NR, and KOG database. Gene ontology annotation and Kyoto Encyclopedia of Genes and Genomes classi fication identi fied numbers of unigenes involved in biomineralization and pigmentation. Digital gene expression analysis revealed that melanin, trace metal elements and porphyrins are potentially involved in shell coloration of A. irradians.Keyword: molluscs; shell coloration; differentially expressed genes; gene ontology
1 INTRODUCTION
The magni ficent color of molluscan shells has attracted the interest of collectors and scientists for centuries (Comfort, 1951) and has served as an important phenotypic marker in shell fish breeding because of its high heritability (Ino, 1949; Leighton,1961; Cole, 1975; Creese and Underwood, 1976;Newkirk, 1980; Palmer, 1985; Adamkewicz and Castagna, 1988; Winkler et al., 2001; Zheng et al.,2013). With the aid of the recently developed nextgeneration sequencing technology, various genes were revealed to be involved in shell coloration of molluscs. For instance, 358 differentially expressed genes are related to color differences in the pearl mussel Hyriopsis cumingii (Bai et al., 2013); the notch signaling pathway and calcium signaling process may equally affect shell coloration in the clam Meretrix meretrix (Yue et al., 2015); a large number of candidate genes in the Japanese scallop Patinopecten yessoensis that are associated with shell biomineralization and metal transcription may be involved in shell coloring (Ding et al., 2015); and in the pearl oyster Pinctada fucata, 10 sequences of 343 randomly selected clones from cDNA subtractive libraries are differentially expressed in white-shelled and red-shelled individuals (Guan et al., 2011).
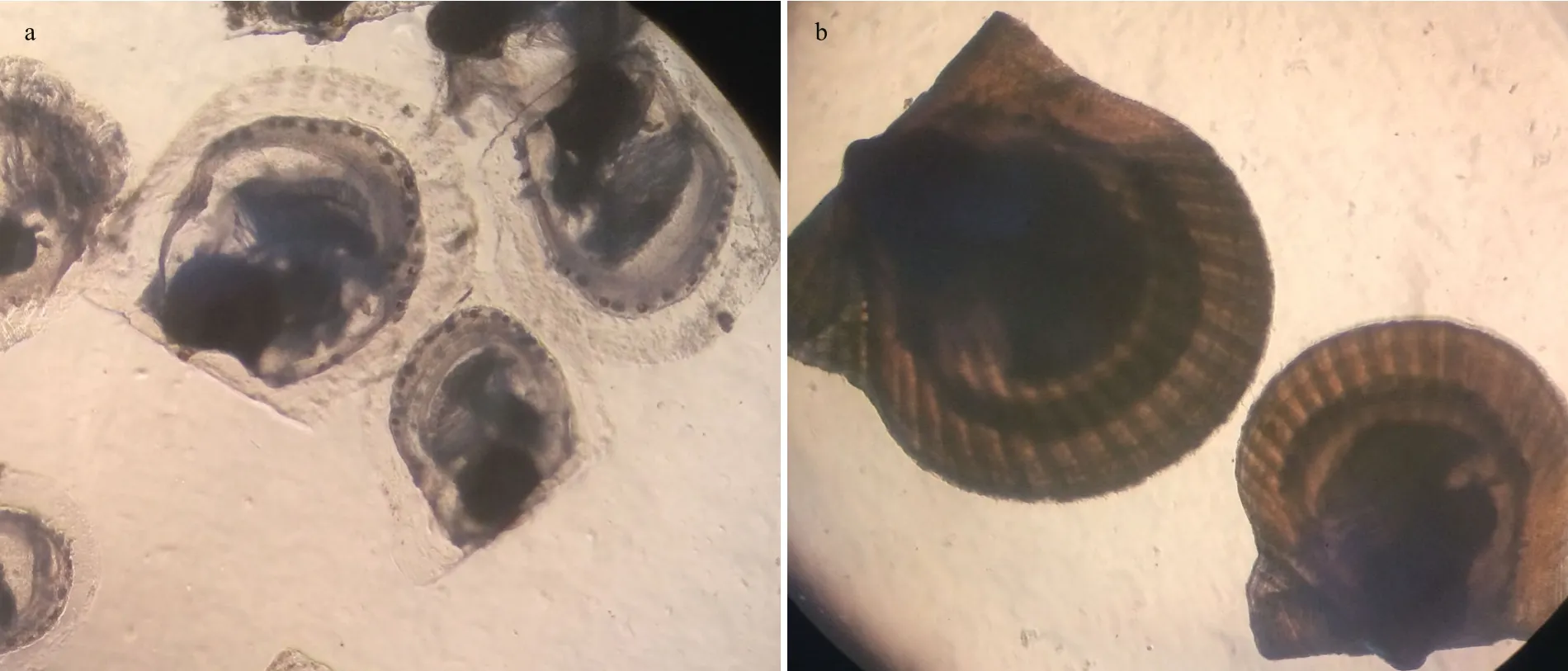
Fig.1 Major transitions during the shell development of A. irradians
The bay scallop A rgopecten irradians (Lamarck 1819) has historically occurred on the coast of the Eastern United States and the Gulf of Mexico. It was introduced to China several times in the 1980s and 1990s (Zhang et al., 1986; Blake et al., 1997; Li et al.,2014) and has been a major cultivated molluscan species for over 20 years. The background color of A. irradians covers the whole shell and never changes throughout its lifetime. It is not only inheritable, but also attributed to a single locus (Adamkewicz and Castagna, 1988; Elek and Adamkewicz, 1990). The orange shell color is one the background colors and has been used as a phenotypic marker in the“zhongkehong” variety (accession number: GS01-004-2006, National Certi fication Committee for Aquatic Varieties of China) genetically improved for growth and survivorship. Although two ampli fied fragment length polymorphisms (AFLPs) were identi fied to be linked to orange shell color in A.irradians (Qin et al., 2007), little is known about the genetic mechanism underlying the orange coloration.In the present research, high-throughput RNA sequencing and digital gene expression (DGE)analysis were used to identify differentially expressed genes associated with the shell pigmentation of a selfbred “zhongkehong” family, aiming to reveal candidate genes involved in orange shell coloration of the bay scallop.
2 MATERIAL AND METHOD
2.1 Sample collection and mRNA extraction
In this study, a self-fertilizing family was established from a parent from the “zhongkehong” variety. Thirtyday-old individuals with transparent shell color and 50-day-old individuals with orange shell color were stored in RNAstore Reagent (Qiagen, Valencia, CA,USA) (Fig.1). Six samples (2 developmental stages ×3 replicates) were pooled in equal weight and macerated in the presence of liquid nitrogen. Given the tiny juveniles of A. irradians, multiple individuals(more than 100) were pooled to generate RNA for each sample. The RNA was isolated from each sample using an RNAprep pure Tissue Kit (Tiangen, Beijing,China) following the manufacturer’s protocol. RNasefree DNase I (Qiagen, USA) was used to remove the residual DNA. An Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and NanoDrop Spectrophotometer (Thermo Scienti fic, Wilmington,DE, USA) were employed to con firm the RNA integrity.
2.2 Library preparation and Illumina HiSeq sequencing
A paired-end cDNA library was prepared using an mRNA-seq sample preparation kit (TruSeq RNA Sample Preparation Kit; Illumina Inc., San Diego,CA, USA). First, mRNA was puri fied using oligo-dT magnetic beads and fragmented by incubation in a fragmentation reagent. Second, random hexamer primers and reverse transcriptase (Invitrogen,Carlsbad, CA, USA) were utilized to synthesize first strand cDNA based on the fragmented mRNA. Then the second strand cDNA was primed mainly using DNA polymerase I (New England BioLabs, Ipswich,MA, USA) and RNase H (Invitrogen, USA). Endrepairing of the double-stranded cDNA was ful filled using T4 DNA polymerase, and the Klenow fragment was added to the 3′ end of the blunt fragment immediately before the index adapter was ligated.Finally, the DNA fragment was enriched by PCR ampli fication and quanti fied in an Aglient 2100 Bioanalyzer before sequencing. The sequencing of the library was performed by Oebiotech (Shanghai,China) on an Illumina HiSeq 2500 platform to obtain paired-end reads at 125 nt.
2.3 De novo transcriptome analysis
Before transcriptome assembly, quality checking of the raw data was performed using FASTQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/).The per base sequence quality plots and the per base sequence content for the paired-end reads from each library indicated good quality of the raw data. To eliminate the error and bias in sequencing data, NGS QC TOOLKIT v2.3.3 (Patel and Jain, 2012) was used for preprocessing, which included removing low quality reads (PHRED score <20), reads with low quality 3′ terminal bases (PHRED score <20), and those with repeated unknown bases (>35). In addition,read contamination was determined for each sample by comparing 500 000 randomly selected reads with the NCBI database (ftp://ftp.ncbi.nih.gov/blast/db);ten reads with the highest score belonged to A. irradians and related species, con firming no contamination. Transcripts of these high quality clean data were obtained using Trinity (Grabherr et al.,2011; Haas et al., 2013). The TIGR Gene Indices clustering tool (TGICL) (Pertea et al., 2003) was used to assemble unigene sequences.
2.4 Annotation of unigene sequences
Using the BLASTx algorithm (Altschul et al.,1990), the assembled contigs were aligned against the GenBank non-redundant (NR) protein database.Furthermore, functional annotation was performed by BLAST comparison against SWISS-PROT (Boutet et al., 2016) and the EuKaryotic Orthologous Groups of proteins (KOG) (Tatusov et al., 2003). The best aligned results were used to infer the sequence direction of unigenes. According to the protein annotation of NR, gene ontology (GO) annotation was performed using Blast2GO (Conesa et al., 2005)with E-value<10-6. KEGG annotation was conducted by software KAAS and KEGG Automatic Annotation Server with E-value<10-10(Moriya et al., 2007).
2.5 Gene expression quanti fication and differential expression analysis
The assembled de novo transcriptome was used as the reference database, and the reads were mapped to the reference transcriptome. This enables us to quantitate gene expression by counting reads per genes.Brie fly, clean data of gene expression quanti fication were mapped back to the reference transcriptome using Bowtie v0.12.9 (Langmead and Salzberg, 2012) and eXpress (Roberts and Pachter, 2012).
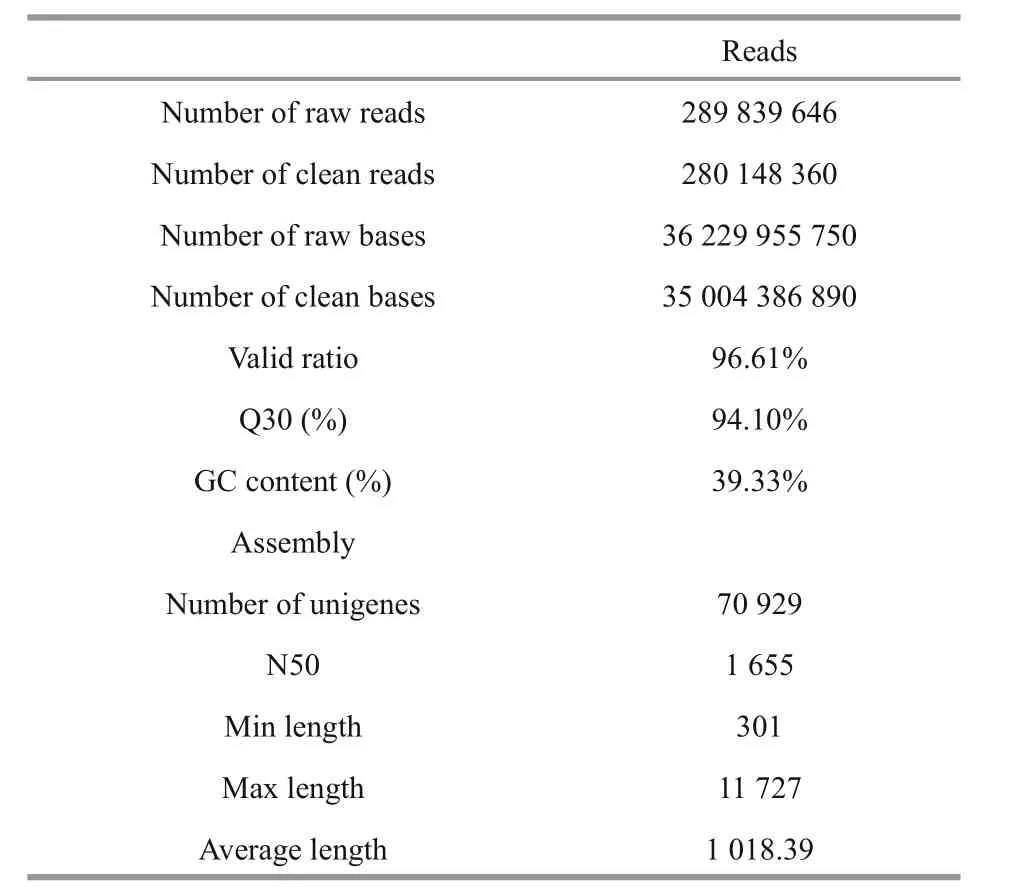
Table 1 Raw reads and assembly statistics for a transcriptome assembly of paired-end reads from two sequencing lanes of an Illumina HiSeq2500
Subsequently, each gene’s FPKM (fragments per kilobase per million mapped reads) was calculated based on the length and read counts. The potential outliers were checked by principal component analysis (PCA) and plotting of the principal components. We performed pair-wise comparisons of the gene expression of the six transcriptomes using the DESeq R package (Anders and Huber, 2010),which employs the negative binomial distribution as the basis of the modeling of RNA-seq counts. Then,MA plots and heatmaps were drawn to assess the transcriptional pattern variations. During pathway and GO enrichment analysis, signi ficantly enriched KEGG and GO terms were identi fied based on threshold of P <0.05.
3 RESULT AND DISCUSSION
3.1 Paired-end sequencing and assembly
The Illumina sequencing of the six cDNA libraries generated 289 839 646 paired-end reads. The per base sequence quality of the paired-end reads was good,and the base distribution examination indicated no sequence-speci fic bias. Subsequently, the original number of sequences was reduced to 280 148 360(96.61%) clean reads after removal of reads with poor quality and unique identi fiers. The clean sequences were assembled into 70 929 transcripts using Trinity software. The average length of final unigenes was 1 018.39 bp ranging from 301 bp to 11 727 bp and the N50 was 1 655 (Table 1).
3.2 Functional annotation of the transcriptome

Fig.2 Top-hit species distribution of A. irradians unigenes against GenBank non-redundant (NR) database
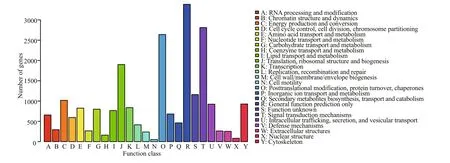
Fig.3 Eukaryotic Orthologous Groups (KOG) function classi fications of A. irradians
Using an E-value cutoff at or below 10-5for BLASTX and BLASTN, the assembled unigenes were annotated against SWISS-PROT, NR, and KOG databases: 23 598 (33.27%), 30 896 (43.56%), and 20 084 (28.32%) unigenes were annotated in SWISSPROT, NR, and KOG database, respectively. The best matched unigenes were selected (Table S1). The tophit species distribution of the unigenes against the NR database is shown in Fig.2. Overall, 44.33% of the hits matched to Crassostrea gigas, whereas the other hits matched to many other species but each accounted for a very small proportion. Eukaryotic Orthologous Groups (KOG) analysis classi fied the 20 084 unigenes into 25 functional categories. The category ‘general function prediction only’ contained the largest proportion of unigenes (3 384; 16.85%), followed by the ‘signal transduction mechanisms’ cluster (2 811;14.00%), and the ‘post-translational modi fication,protein turnover, chaperones’ cluster (2 640; 13.14%)(Fig.3).
3.3 Gene ontology (GO) classi fication
The GO analysis of 30 896 unigenes annotated in the NR database was conducted with Blast2GO. In total, 21 755 unigenes were assigned to three main GO categories with 12 924 functional terms and further subdivided into 64 subcategories (Fig.4).Among these GO term assignments, 66.15% were from the biological processes category, 23.5% were from the molecular function category, and 10.35%were from the cellular component category. Twelve annotated unigenes were involved in biomineralization and 52 annotated unigenes were involved in pigmentation (Table S2).
3.4 Kyoto Encyclopedia of Genes and Genomes(KEGG) classi fication
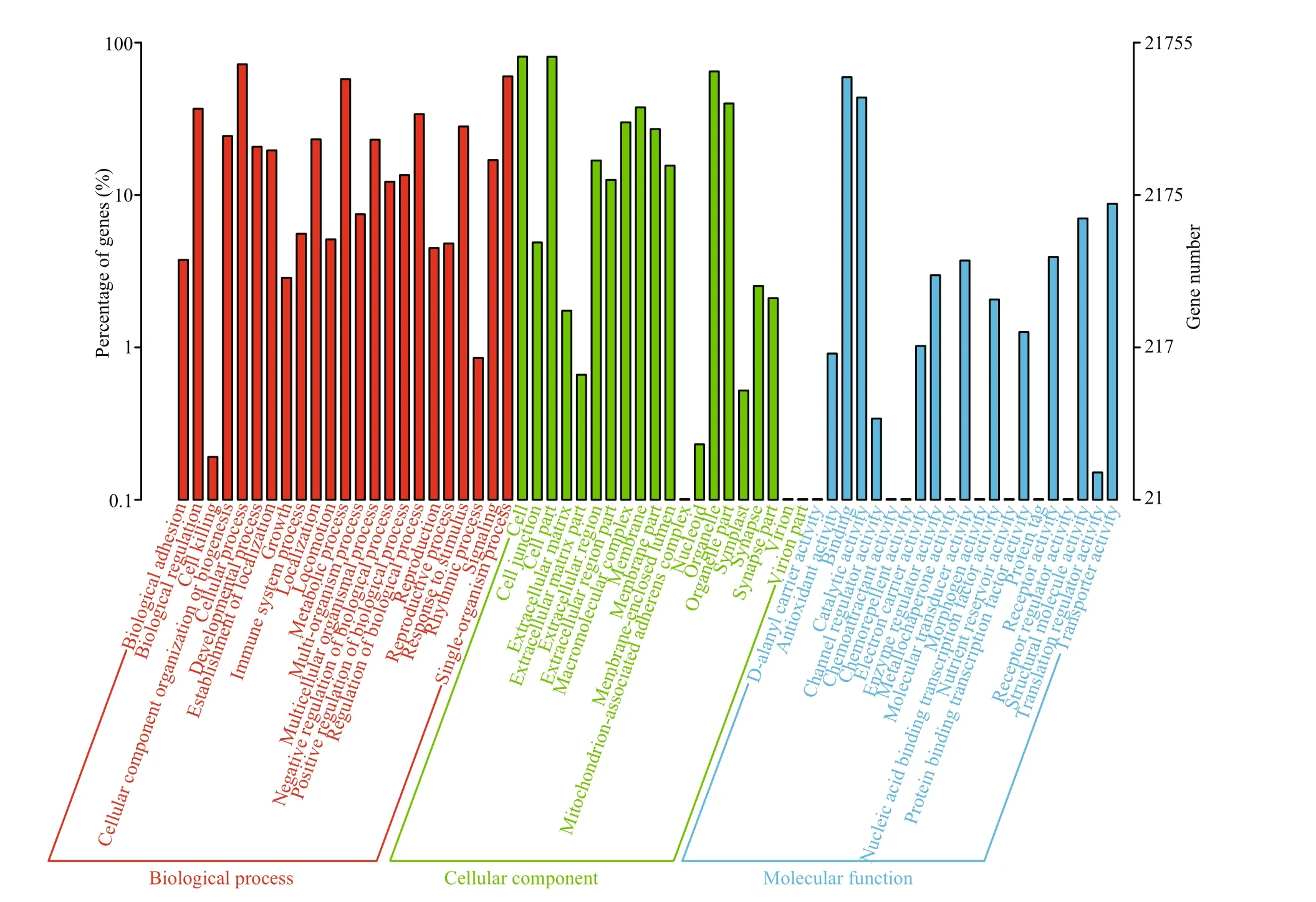
Fig.4 Gene ontology annotation results of assembled unigenes
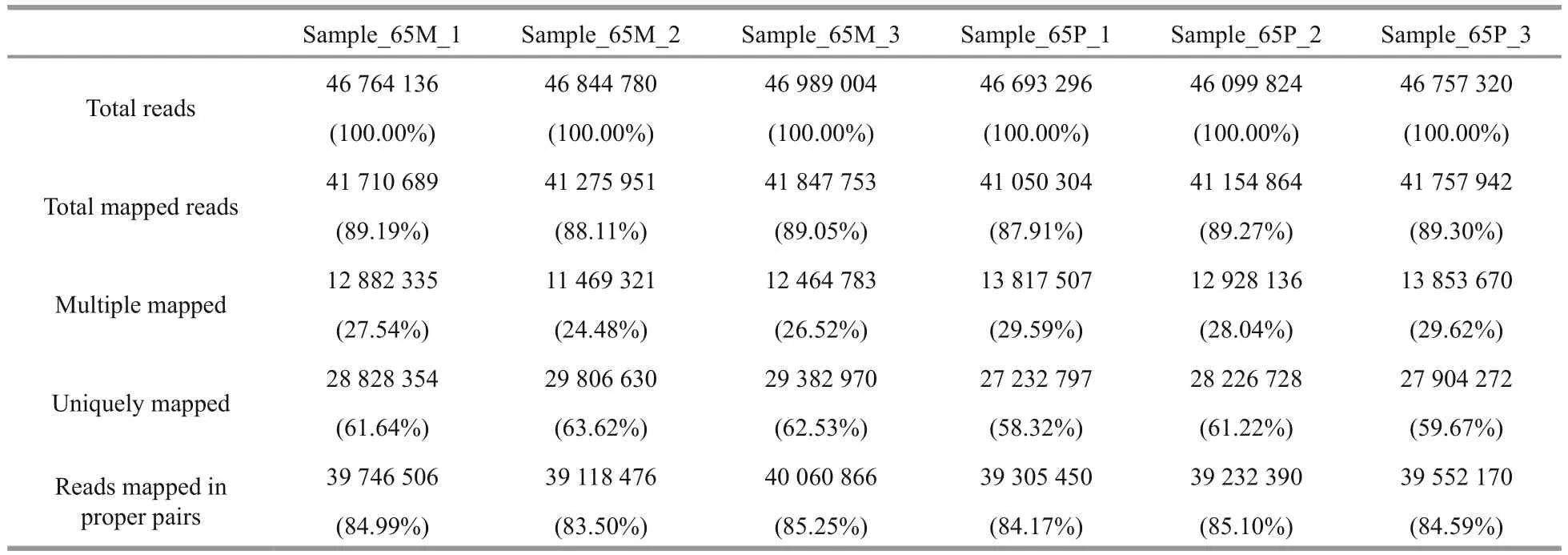
Table 2 Mapping statistics of reads to the unigenes
In total, 11 347 unigenes were assigned to 350 KEGG pathways. Of these pathways, 25.14% were included in metabolism, and three major subgroups were involved in lipid metabolism, carbohydrate metabolism, and xenobiotics biodegradation and metabolism. Other pathways included organismal systems (21.71%), human diseases (20.86%),environmental information processing (10.00%),genetic information processing (6.29%), and cellular processes (6.00%).
3.5 Gene expression quanti fication and comparative analysis
All the unigenes from the six transcriptome libraries were used as the reference transcriptome for reads mapping. The mapping rates exceeded 87% for total mapped, 58% for uniquely mapped, and 83% for reads mapped in proper pairs (Table 2).
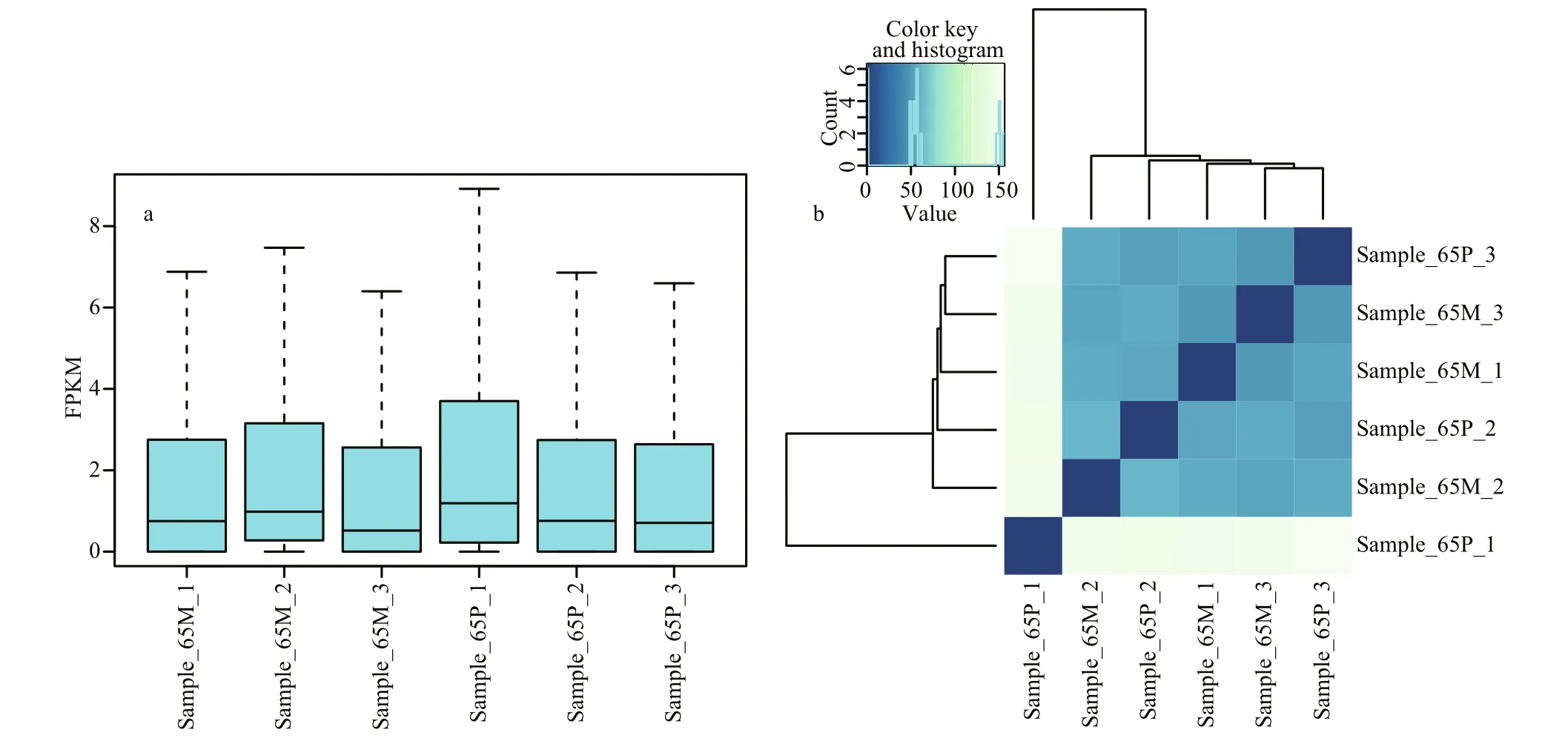
Fig.5 Transcripts abundance and sample-to-sample analysis based on the expressive quantities of the genes
Based on the FPKM (Mortazavi et al., 2008), the transcript abundance of the six libraries was calculated and a box-whisker plot was drawn (Fig.5a). The PCA and sample-to-sample cluster analysis based on the expressive quantities of these genes (Fig.5b) identi fied sample_65P_1 as an outlier. This may be due to sampling error (individuals collected in this sample were getting shell colored), and the outliers were removed from following analysis.
Differential expression (DE) analysis was performed using the DESeq R package to screen for differentially expressed genes (DEGs) ( P ≤ 0.05).Consequently, 1 486 DEGs were identi fied, of which 748 were up-regulated and 738 were down-regulated(Table S3). The MA plot visually summarizes the obtained results (Fig.6a). The global expression pro files of the DEG union in each DGE library were estimated by hierarchical clustering (Fig.6b); the results showed that the global expression pattern of DEGs in the case group (post-pigmentation) was distinguishable from that in the control group (prepigmentation).
3.6 Functional analysis of differentially expressed genes
Using a reference gene database (30 896 annotated unigenes), 675 DEGs were successfully annotated and subjected to GO and KEGG pathway enrichment analysis to better delineate their function. In total, 453 DEGs were assigned to at least one GO term and 266 DEGs were annotated in the KEGG pathway database.Melanogenesis (ko04916, P <0.05), phenylalanine tyrosine and tryptophan biosynthesis (ko00400,P <0.01), tyrosine metabolism (ko00350, P <0.01),iron ion transport (GO:0006826, P <0.05), and porphyrin and chlorophyll metabolism (ko00860,P <0.05) were signi ficantly enriched in the upregulated genes.
Melanins are the group of black, brown, and reddish materials derived from tyrosine (Arnow,1938). In vertebrates, the red and yellow color of hairs and feathers is largely due to pheomelanins(Zviak and Milléquant, 2005). The ink of coleoid cephalopods is blackened by melanin (Derby, 2014),and the pigment in the adductor muscle scar of the Paci fic oyster is a melanin (Hao et al., 2015). In the present study, melanogenesis was highly enriched in the DEGs of the orange shell. Additionally,phenylalanine tyrosine and tryptophan biosynthesis and tyrosine metabolism were also recognized, all of which are highly involved in melanin metabolism in mammals (Sánchez-Ferrer et al., 1995). Furthermore,Tyrosinase-like protein 1 (TRP1) was signi ficantly up-regulated in A. irradians, and the melanogenic function of TRP1 had been con firmed to be the oxidation of 5,6-dihydroxyindole-2-carboxylic acid(DHICA) to a carboxylated indole-quinone at a downstream point in the melanin biosynthetic pathway(Kobayashi et al., 1994). Whether melanogenesis,phenylalanine tyrosine and tryptophan biosynthesis,tyrosine metabolism were involved in the shell coloration of A. irradians is an interesting question and further studies are required to provide evidence.
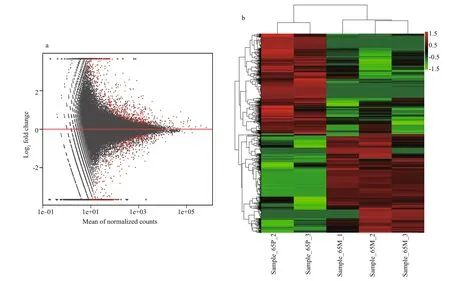
Fig.6 Global expression pro files of differentially expressed genes (DEGs)
Porphyrins are natural and ubiquitous tetrapyrrole avian eggshell pigments. Uroporphyrin I was found to be widely distributed in the shell of pteriomorph bivalves and other gastropod clades (Comfort, 1951;Creese & Underwood, 1976; Fox, 1976). These biological pigments were reported to be associated with red, brown or purple shell colouration (Comfort,1949). In recent studies, the shell of Argopecten sp.,one of the close relatives of A. irradians, was determined to contain small quantities of protoporphyrin using high-performance liquid chromatography (HPLC) (Verdes et al., 2015).Moreover, trace metals were reported to bind to porphyrins to form metalloporphyrin and metalloporphyrin, where different trace metals would result in differing coloration (Williams et al., 2016). It had been reported that trace metals, especially Fe2+and Zn2+, and metal transcription are linked closely to shell coloring of P. yessoensis (Ding et al., 2015). In the present study, zinc ion binding (GO: 0008270),copper ion binding (GO: 0006825), and porphyrin and chlorophyll metabolism (ko00860) were also enriched in DEGs. This implicates that these trace metals and porphyrins were involved in the progress of A. irradians orange shell pigmentation; further study is required.
4 CONCLUSION
The de novo transcriptome of A. irradians from two developmental stages, pre-pigmentation and post-pigmentation, was determined using the Illumina platform. A total of 70 929 unigenes were obtained and 30 896 unigenes were successfully annotated,which provided more sequences and genetic information to A. irradians, for which there is no reference genome available. According to GO analysis and KEGG database annotation, various unigenes were involved in biomineralization and pigmentation. Functional analysis of DEGs suggested melanins, trace metal elements and porphyrins may be potentially involved in the shell coloration of the bay scallop A. irradians. To support these finding,qRT-PCR should be employed to test pigmentation related genes in a future study.
6 ACKNOWLEDGMENT
Thanks are due to Laizhou Shunchang Aquatic Products Co., Ltd. for assistance with the experiments and to Mr. DU Runshan and Mr. YANG Linying for sample collection. In addition, we are grateful to Dr.WANG Jinpeng and Dr. Song Kai for their suggestions on the data analysis.
5 DATA AVAILABILITY STATEMENT
The raw reads generated during the current study are available in the NCBI SRA database, the accession number is SRR5469239.
 Journal of Oceanology and Limnology2018年4期
Journal of Oceanology and Limnology2018年4期
- Journal of Oceanology and Limnology的其它文章
- Editorial Statement
- Effects of seawater acidi fication on the early development of sea urchin Glyptocidaris crenularis*
- Dietary effects of A zolla pinnata combined with exogenous digestive enzyme (Digestin?) on growth and nutrients utilization of freshwater prawn, Macrobrachium rosenbergii(de Man 1879)
- Preliminarily study on the maximum handling size, prey size and species selectivity of growth hormone transgenic and non-transgenic common carp Cyprinus carpio when foraging on gastropods*
- Hydrodynamic characteristics of the double-winged otter board in the deep waters of the Mauritanian Sea*
- New record of Japanese snake blenny Xiphasia matsubarai(Perciformes: Blenniidae) from South China Sea*
